Research
We work on developing and applying new statistical and computational methods that use genetic data for better understanding how populations and species evolve. Our goal is to develop new perspectives, new tools and new ideas to interpret genetic data.
In the past years, we have been heavily focused on genetic data retrieved from fossils (ancient DNA), particularly from early modern humans, Neandertals and Denisovans. These data give us unprecedented insights into how our ancestors lived, interacted and evolved.
Population Genetics of Early Modern Humans, Neandertals and Denisovans
 Human and Hominin Ancient DNA has recently provided a new way into how we can
study our distant past. Under ideal conditions, DNA can survive tens to hundreds
of thousands of years, and gives us a glimpse into populations from the past.
Human and Hominin Ancient DNA has recently provided a new way into how we can
study our distant past. Under ideal conditions, DNA can survive tens to hundreds
of thousands of years, and gives us a glimpse into populations from the past.
In collaboration with the Department of Evolutionary Genetics at the MPI for Evolutionary Anthropology, group members contributed to several studies that published data from Neandertals and early modern humans. For example, we led the first analysis of the genetic data from an entire Neandertal community. This study, lead by postdoc Laurits Skov, sequenced 13 new Neandertals from Chagyrskaya Cave (Altai mountains, Russia). In it, we showed that the Neandertals from Chagyrskaya Cave all lived at the same time, lived in very small communities, and that they were mainly connected through female migration. This work has been widely covered in print and online media.
Selected Publications
- Skov L, Peyrégne S, Popli D, Iasi LNM, Devièse T, Slon V, Zavala EI, Hajdinjak M, Sümer AP, Grote S, Bossoms Mesa A, López Herráez D, Nickel B, Nagel S, Richter J, Essel E, Gansauge M, Schmidt A, Korlević P, Comeskey D, Derevianko AP, Kharevich A, Markin SV, Talamo S, Douka K, Krajcarz MT, Roberts RG, Higham T, Viola B, Krivoshapkin AI, Kolobova KA, Kelso J, Meyer M, Pääbo S Peter, BM. Genetic insights into the social organization of Neanderthals Nature
- Vernot B, Zavala EI, Gómez-Olivencia A, Jacobs Z, Slon V, Mafessoni F, Romagné F, Pearson A, Petr M, Sala N, Pablos A, Aranburu A, Bermúdez de Castro JM, Carbonell E, Li B, Krajcarz MT, Krivoshapkin AI, Kolobova KA, Kozlikin MB, Shunkov MV, Derevianko AP, Viola B, Grote S, Essel E, López Herráez D, Nagel S, Nickel B, Richter J, Schmidt A, Peter BM, Kelso J, Roberts RG, Arsuaga JL, Meyer M: Unearthing Neanderthal population history using nuclear and mitochondrial DNA from cave sediments. Science
- Huerta-Sánchez E, Jin X, Bianba Z, Peter BM et al. (2014): Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature
Methods for the analysis of Ancient DNA
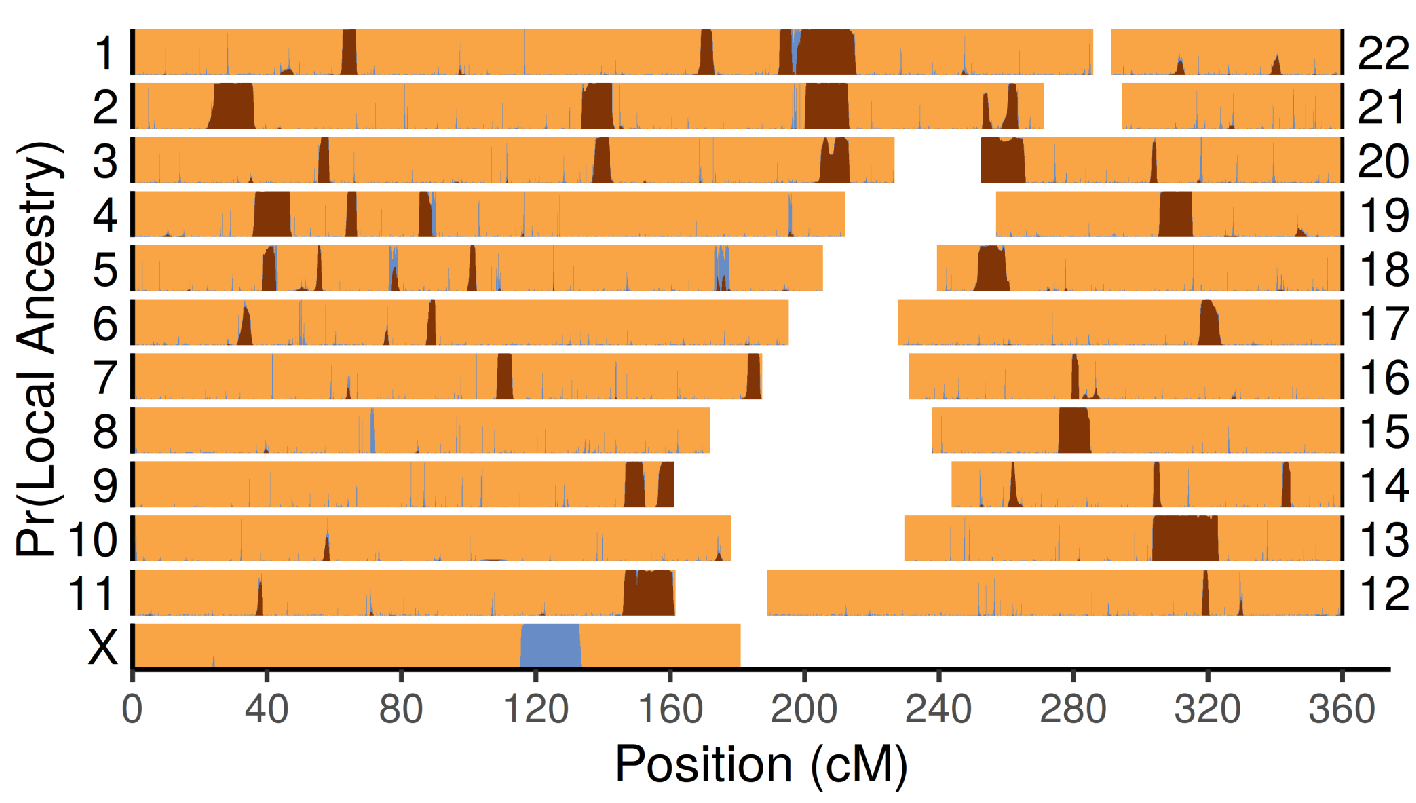 Much of our contribution in the analysis of ancient DNA is in developing new
population genetic methods. One of the major challenges in ancient DNA is the
rapid increase in data; since the publication of the draft Neandertal genome in
2010, the field now produces genetic data from thousands of individuals each year.
Our methods aim to leverage this wealth of information, while tackling the core
challenges pertaining to ancient DNA
Much of our contribution in the analysis of ancient DNA is in developing new
population genetic methods. One of the major challenges in ancient DNA is the
rapid increase in data; since the publication of the draft Neandertal genome in
2010, the field now produces genetic data from thousands of individuals each year.
Our methods aim to leverage this wealth of information, while tackling the core
challenges pertaining to ancient DNA
- ancient genomes are sampled at different time points throughout history
- ancient DNA is often extremely sparse, low quality and contaminated
- we now have genetic information from hundreds to thousands of individuals we would like to analyze jointly.
For example, we recently published a new method, KIN, to infer familial relatedness from low-coverage ancient DNA data. Another recent example of our work is the program admixfrog, that is able to estimate local ancestry from low-coverage, contaminated populations. Using it, we showed that Early East Asians that lived more than 30’000 years ago already had Denisovan ancestry, and that some of the earliest modern humans in Europe, from Bacho Kiro in Bulgaria, had very recent Neandertal ancestry.
Selected Publications
- Popli D, Peyrégne S & Peter, BM: KIN: A method to infer relatedness from low-coverage ancient DNA Genome Biology
- Hajdinjak M, Mafessoni F, Skov L, Vernot B, Hubner A, Fu Q, Essel E, Nagel S, Nickel B, Richter J, Moldovan OT, Constation S,, Endarova E, Zahariev N, Spasov R, Welker F, Smitt GM, Sinet-Mathiot V, Paskulin L, Fewlass H, Talamo S, Rezek Z, Sirakova S, Sirakov N, McPherron SP, Tsanova T, Hublin JJ, Peter BM, Meyer M, Skoglund P, Kelso J, Paabo S: Initial Upper Palaeolithic humans in Europe had recent Neanderthal ancestry. Nature
- Massilani D, Skov L(http://dx.doi.org/10.7554/eLife.59323), Hajdinjak M, Gunchinsuren B, Tseveendorj D, Yi S, Lee J, Nagel S, Nickel B, Devies T, Higham T, Meyer M, Kelso J, Peter BM, Paabo S: Denisovan ancestry and population history of early East Asians. Science
Genetic Diversity Through Space and Time
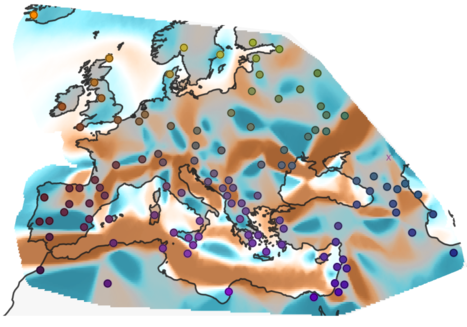 We now have genetic information from hundreds to thousands of people. Ideally,
we would like to analyze them jointly, but this is often difficult to
conceptualize, model and understand.
We now have genetic information from hundreds to thousands of people. Ideally,
we would like to analyze them jointly, but this is often difficult to
conceptualize, model and understand.
Publications
- Iasi, LNM, Ringbauer H & Peter BM: An extended admixture pulse model reveals the limitations to Human-Neandertal introgression dating. (Molecular Biology and Evolution)
- Peter, BM, Petkova D , Novembre J: Genetic landscapes reveal how human genetic diversity aligns with geography. Molecular Biology and Evolution
- Peter BM and Slatkin M (2015): The effective founder effect in a spatially expanding population. Evolution
- Peter BM and Slatkin M (2013): Detecting Range Expansions from Genetic Data. Evolution
- Peter BM, Wegmann D, and Excoffier L (2010): Distinguishing between population bottleneck and population subdivision by a Bayesian model choice procedure. Molecular Ecology
F-Statistics
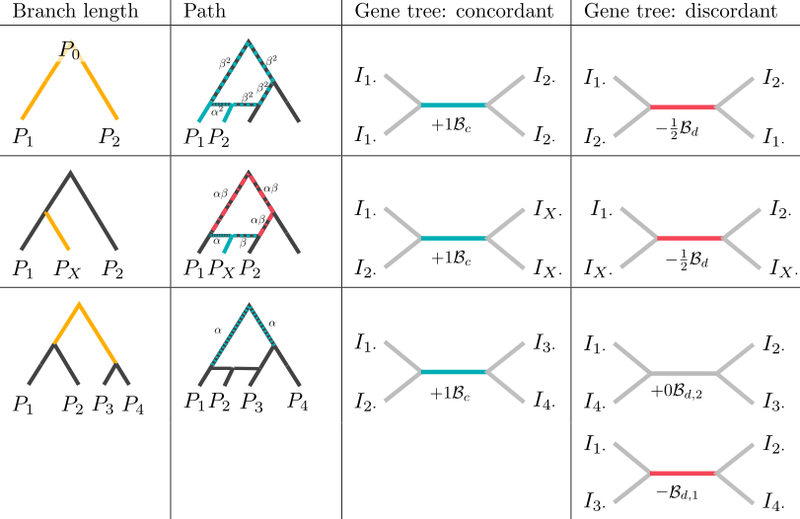 F-statistics are a set of statistics that are widely used for the analysis of
population structure, particularly in ancient DNA. Our work focuses on better
understanding these statistics.
F-statistics are a set of statistics that are widely used for the analysis of
population structure, particularly in ancient DNA. Our work focuses on better
understanding these statistics.
Publications
- Peter BM: A geometric relationship of F2, F3 and F4-statistics with principal component analysis. Philosophical Transactions of the Royal Society B: Biological Sciences
-
Peter BM (2016): Admixture, Population Structure, and F-Statistics. Genetics 202:1485-1501; .